Sharpness is the ability of the model to generate predictions within a narrow range and dispersion is the lack thereof. It is a data-independent measure, and is purely a feature of the forecasts themselves.
Dispersion of predictive samples corresponding to one single observed value is measured as the normalised median of the absolute deviation from the median of the predictive samples. For details, see mad() and the explanations given in Funk et al. (2019)
Arguments
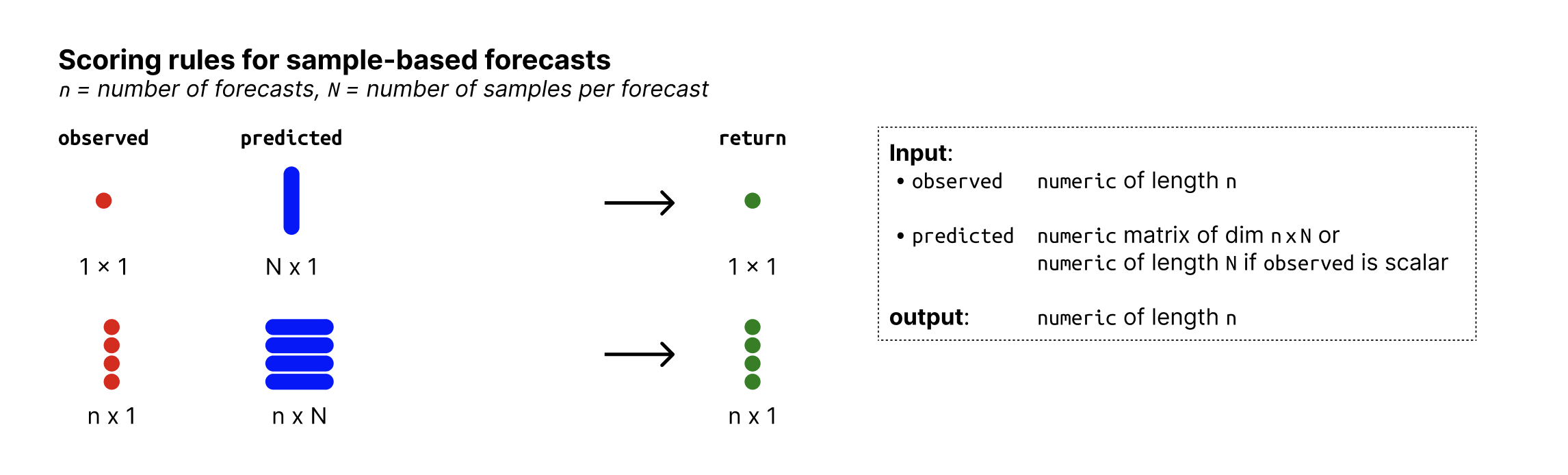
- observed
Place holder, argument will be ignored and exists only for consistency with other scoring functions. The output does not depend on any observed values.
- predicted
nxN matrix of predictive samples, n (number of rows) being the number of data points and N (number of columns) the number of Monte Carlo samples. Alternatively, if n = 1,
predictedcan just be a vector of size n.- ...
Additional arguments passed to mad().
References
Funk S, Camacho A, Kucharski AJ, Lowe R, Eggo RM, Edmunds WJ (2019) Assessing the performance of real-time epidemic forecasts: A case study of Ebola in the Western Area region of Sierra Leone, 2014-15. PLoS Comput Biol 15(2): e1006785. doi:10.1371/journal.pcbi.1006785
Examples
predicted <- replicate(200, rpois(n = 30, lambda = 1:30))
mad_sample(predicted = predicted)
#> [1] 1.4826 1.4826 1.4826 1.4826 1.4826 2.9652 2.9652 2.9652 2.9652 2.9652
#> [11] 3.7065 2.9652 2.9652 4.4478 3.7065 4.4478 4.4478 2.9652 2.9652 4.4478
#> [21] 4.4478 4.4478 4.4478 4.4478 5.9304 5.9304 4.4478 5.9304 5.9304 5.9304