Returns key summary plots for estimates. May be depreciated in later
releases as current S3 methods are enhanced.
Arguments
- summarised_estimates
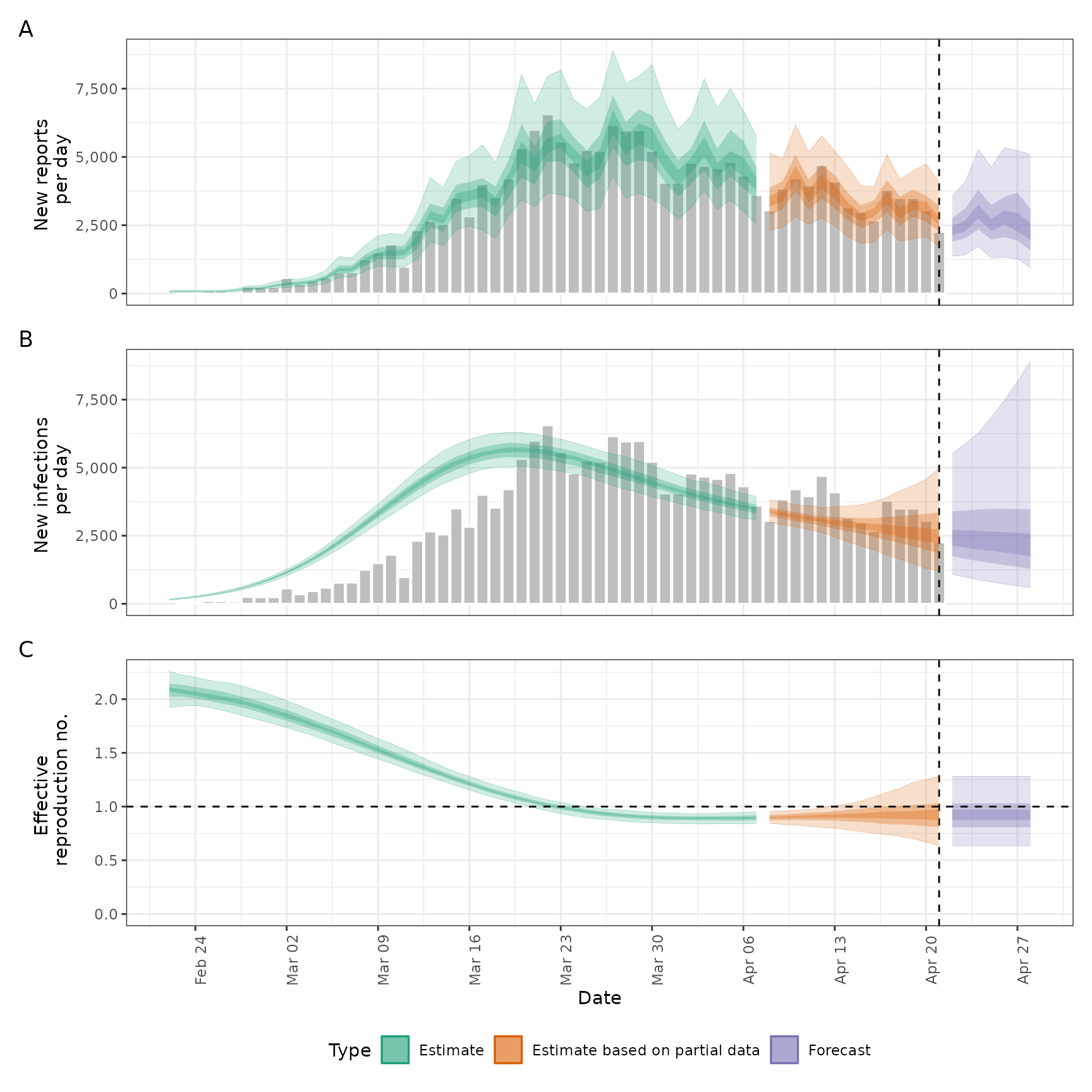
A data.table of summarised estimates containing the following variables: variable, median, bottom, and top.
It should also contain the following estimates: R, infections, reported_cases_rt, and r (rate of growth).
- reported
A
<data.table>of reported cases with the following variables: date, confirm.- target_folder
Character string specifying where to save results (will create if not present).
- ...
Additional arguments passed to
plot_estimates().
Value
A named list of ggplot2 objects, list(infections, reports, R, growth_rate, summary), which correspond to a summary combination (last
item) and for the leading items.
See also
summarised_estimates[variable == "infections"],
summarised_estimates[variable == "reported_cases"],
summarised_estimates[variable == "R"], and
summarised_estimates[variable == "growth_rate"], respectively.
Examples
# get example output form estimate_infections
out <- readRDS(system.file(
package = "EpiNow2", "extdata", "example_estimate_infections.rds"
))
# plot infections
plots <- report_plots(
summarised_estimates = summary(out, type = "parameters"),
reported = out$observations
)
plots
#> $infections
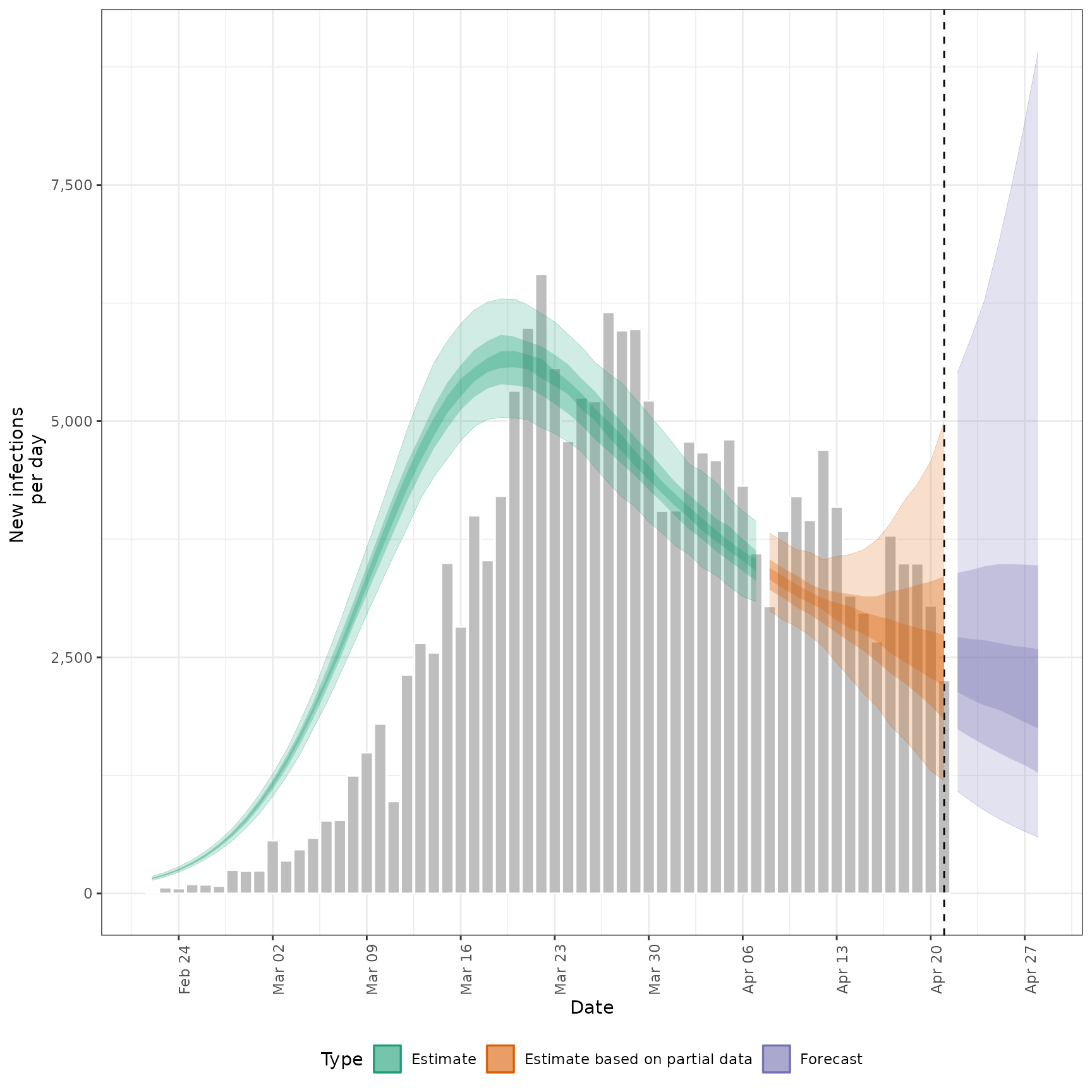 #>
#> $reports
#>
#> $reports
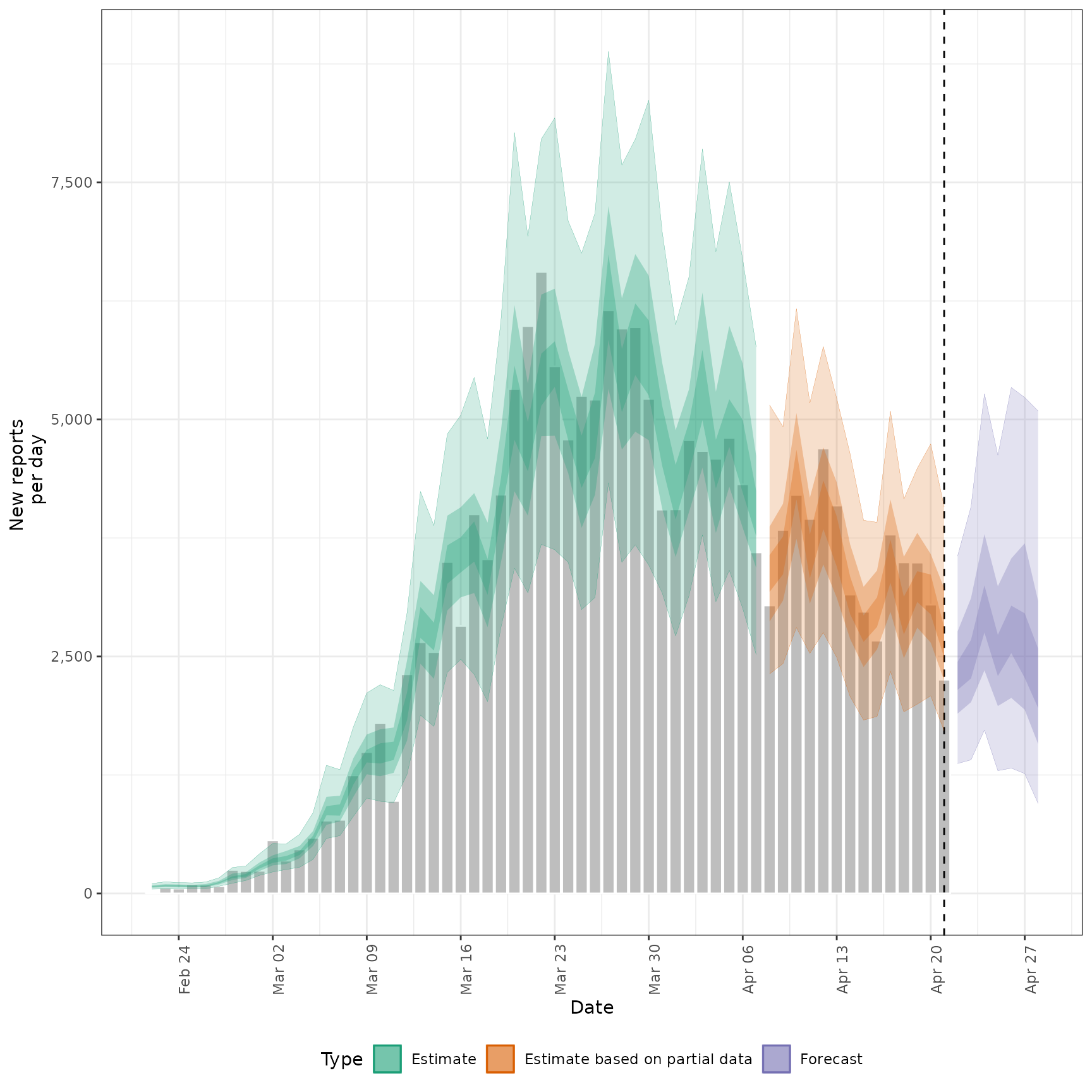 #>
#> $R
#>
#> $R
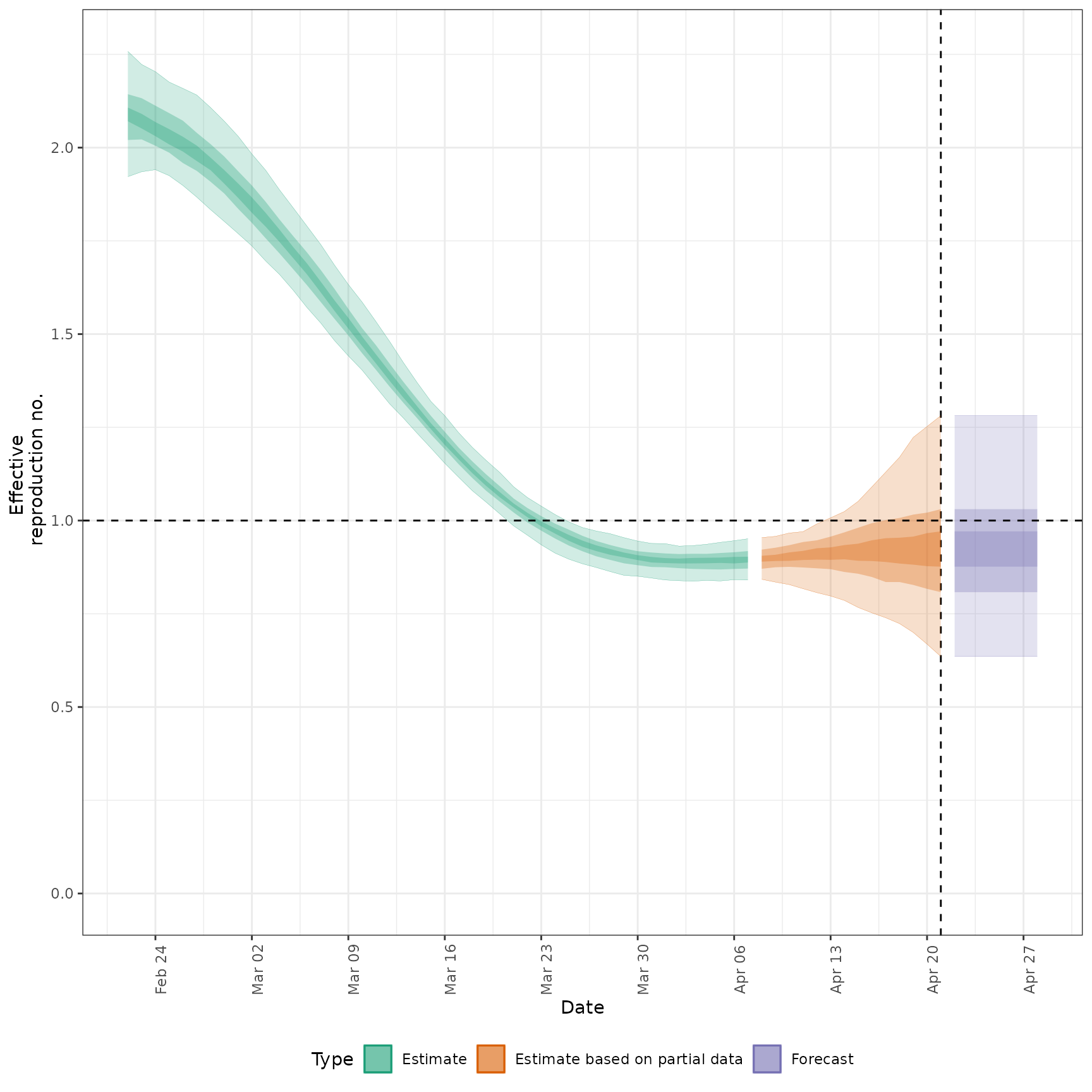 #>
#> $growth_rate
#>
#> $growth_rate
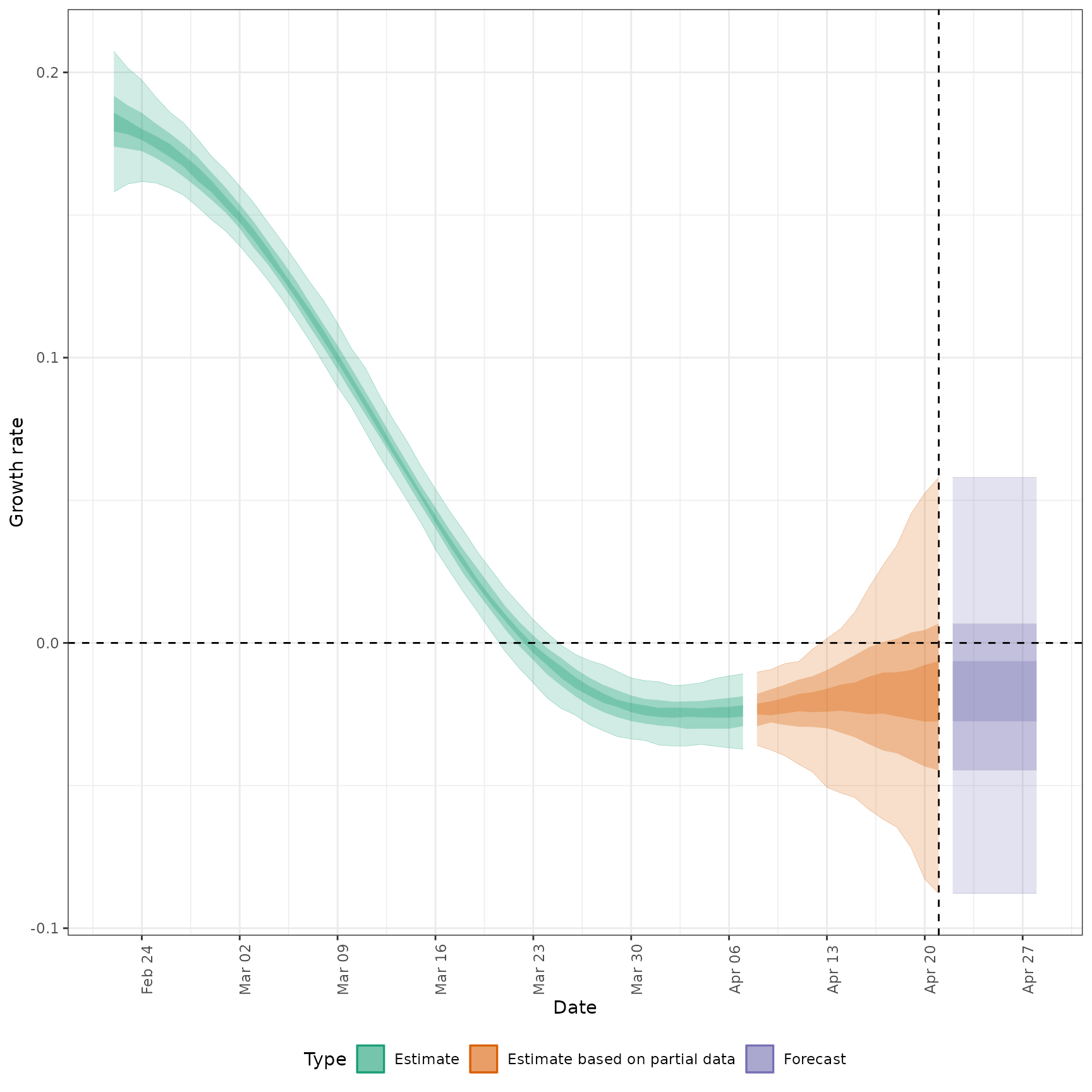 #>
#> $summary
#>
#> $summary
 #>
#>
